Molecular Biomedicine | Interplay between SMAD2 and STAT5A is a critical determinant of IL-17A/IL-17F differential expression

Open the phone and scan
Interleukins (IL)-17A and F are critical cytokines in anti-microbial immunity but also contribute to auto-immune pathologies. Recent evidence suggests that they may be differentially produced by T-helper (Th) cells, but the underlying mechanisms remain unknown. To address this question, we built a regulatory graph integrating all reported upstream regulators of IL-17A and F, completed by ChIP-seq data analyses. The resulting regulatory graph encompasses 82 components and 136 regulatory links. The graph was then supplemented by logical rules calibrated with original flow cytometry data using naive CD4+ T cells, in conditions inducing IL-17A or IL-17F. The model displays specific stable states corresponding to virtual phenotypes explaining IL-17A and IL-17F differential regulation across eight cytokine stimulatory conditions. Authors' model analysis points to the transcription factors NFAT2A, STAT5A and SMAD2 as key regulators of the differential expression of IL-17A and IL-17F, with STAT5A controlling IL-17F expression, and an interplay of NFAT2A, STAT5A and SMAD2 controlling IL-17A expression. We experimentally observed that the production of IL-17A was correlated with an increase of SMAD2 transcription, and the expression of IL-17F correlated with an increase of BLIMP-1 transcription, together with an increase of STAT5A expression (mRNA), as predicted by our model. Interestingly, RORγt presumably plays a more determinant role in IL-17A expression as compared to IL-17F expression. In conclusion, authors propose the first mechanistic model accounting for the differential expression of IL-17A and F in Th cells, providing a basis to design novel therapeutic interventions in auto-immune and inflammatory diseases.

Authors first conducted an extensive analysis of published data to identify the key signaling events and molecules driving Th17 differentiation upon exposure to antigen and various cytokine combinations, including IL-1β, IL-23, IL-6 and TGF-β. They also curated the literature for the IL-12 pathway in T cells in order to understand its interplay with known Th17 inducers.
The IL-12 module includes the input cytokine IL-12, which can trigger the activation of both STAT1 and STAT4, leading to the expression of the master transcriptional regulator T-bet and to the production of IFN-γ cytokine (IL-12 module, Fig. 1). It is usually considered that Th1 and Th17 differentiation programs are mutually exclusive, with mutual inhibitions between T-bet and RORγt. However, based on their literature search, they were led to consider additional cross-talks, in particular an activation of IL-17F by STAT1 and an activation of STAT1 by IL6R.
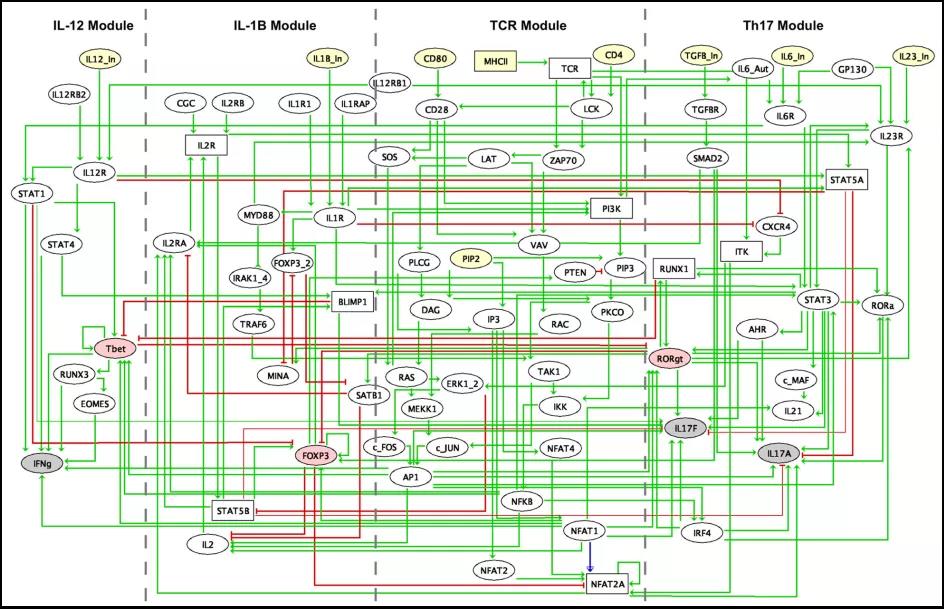
Fig.1 Regulatory graph integrating the interactions inferred from the literature, databases and ChIP-seq data meta-analyses
Article Access: https://link.springer.com/article/10.1186/s43556-021-00034-3
Website for Molecular Biomedicine: https://www.springer.com/journal/43556
Looking forward to your contributions


